PyData.Tokyo Meetup #21 LightGBM / Optuna に参加してから1ヶ月程経ってしまいましたが, Optuna に入門しました。 pfnet/optuna 内の LightGBM の example を実行したのでインストールや使い方を備忘録として残しておきます。
環境は macOS Mojave (10.14.2), Python 3.7.4 です。
Install Optuna
Optuna はハイパーパラメータの最適化ツールで, その機能は既に多くの記事で紹介されている。大きく以下の3つの特徴がある。
- Define-by-Run
- Parallel distributed optimization
- Pruning of unpromising trials
サンプリング法や ASHA (Asynchronous Successive Halving Algorithm) などの技術的な詳細や既存の最適化ツールとの比較は [1] を参照。応用例として FFmpeg の符号化パラメータのチューニングについて紹介されており面白かった。
Optuna を pip で PyPI からインストールする。
$ pip install optuna
Install LightGBM
GBDT (Gradient Boosting Decision Trees) ライブラリである LightGBM をソースからインストールする。具体的には Installation Guide の macOS に記載の手順に沿ってインストールする。
$ brew update
$ brew install cmake
$ brew install libomp
$ git clone --recursive https://github.com/microsoft/LightGBM ; cd LightGBM
$ mkdir build ; cd build
$ cmake \
-DOpenMP_C_FLAGS="-Xpreprocessor -fopenmp -I$(brew --prefix libomp)/include" \
-DOpenMP_C_LIB_NAMES="omp" \
-DOpenMP_CXX_FLAGS="-Xpreprocessor -fopenmp -I$(brew --prefix libomp)/include" \
-DOpenMP_CXX_LIB_NAMES="omp" \
-DOpenMP_omp_LIBRARY=$(brew --prefix libomp)/lib/libomp.dylib \
..
-- The C compiler identification is AppleClang 10.0.0.10001044
-- The CXX compiler identification is AppleClang 10.0.0.10001044
-- Check for working C compiler: /Library/Developer/CommandLineTools/usr/bin/cc
-- Check for working C compiler: /Library/Developer/CommandLineTools/usr/bin/cc -- works
-- Detecting C compiler ABI info
-- Detecting C compiler ABI info - done
-- Detecting C compile features
-- Detecting C compile features - done
-- Check for working CXX compiler: /Library/Developer/CommandLineTools/usr/bin/c++
-- Check for working CXX compiler: /Library/Developer/CommandLineTools/usr/bin/c++ -- works
-- Detecting CXX compiler ABI info
-- Detecting CXX compiler ABI info - done
-- Detecting CXX compile features
-- Detecting CXX compile features - done
-- Found OpenMP_C: -Xpreprocessor -fopenmp -I/usr/local/opt/libomp/include (found version "3.1")
-- Found OpenMP_CXX: -Xpreprocessor -fopenmp -I/usr/local/opt/libomp/include (found version "3.1")
-- Found OpenMP: TRUE (found version "3.1")
-- Configuring done
-- Generating done
-- Build files have been written to: /your/env/LightGBM/build
$ cd ../python-package/
$ python setup.py install
インストールした Optuna と LightGBM を確認する。
$ ipython
Python 3.7.4 (default, Aug 13 2019, 15:17:50)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.8.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]: import lightgbm as lgb
In [2]: import optuna
In [3]: lgb.__version__
Out[3]: '2.3.1'
In [4]: optuna.__version__
Out[4]: '0.17.0'
In [5]: import numpy as np
In [6]: import sklearn.datasets
In [7]: import sklearn.metrics
In [8]: from sklearn.model_selection import train_test_split
Run Examples
今回は pfnet/optuna に含まれる2つの Example (examples/lightgbm_simple.py, examples/pruning/lightgbm_integration.py) を実行する。
Breast Cancer Wisconsin (Diagnostic) dataset
Examples で使われている Breast Cancer Wisconsin (Diagnostic) dataset は1995年に Dr. William H. らによって公開された乳がんの診断データセットで, 現在では二値分類のデータセットとして主に学習やチュートリアルの用途で利用されている。
目的変数は Malignant (悪性), Benign (良性) の二値で 569 サンプルの内, 悪性が 212 サンプル, 良性が 357 サンプルである。
各サンプルは腫瘤に関する以下の10個の各属性について, 平均 (1-10列), 標準誤差 (11-20列), worst または largest (上位3つの値の平均) (21-30列) を計算した計30個の特徴を持つ。
- radius (mean of distances from center to points on the
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area – 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension (“coastline approximation” – 1)
sklearn.datasets からデータセットを読み込む。
In [9]: data, target = sklearn.datasets.load_breast_cancer(return_X_y=True)
In [10]: print(data.shape, target.shape)
(569, 30) (569,)
In [11]: target.sum()
Out[11]: 357
Simple tuning
pfnet/optuna の examples/lightgbm_simple.py を実行する。チューニングする LightGBM のパラメータは以下。
- lambda_l1 (reg_alpha): L1正則化
- lambda_l2 (reg_lambda): L2正則化
- num_leaves: 木の葉の数
- feature_fraction (sub_feature, colsample_bytree): ランダムに選択される特徴の内, 何割の特徴を使うか
- bagging_fraction (bagging): resampling を除いてデータの何割を使うか
- bagging_freq (subsample_freq): バギングを行う間隔 (0 の場合, バギングしない)
- min_data_in_leaf (min_child_samples): ひとつの葉の最小サンプルサイズ
In [12]: def objective(trial):
...: data, target = sklearn.datasets.load_breast_cancer(return_X_y=True)
...: train_x, test_x, train_y, test_y = train_test_split(data, target, test_size=0.25)
...: dtrain = lgb.Dataset(train_x, label=train_y)
...:
...: param = {
...: 'objective': 'binary',
...: 'metric': 'binary_logloss',
...: 'verbosity': -1,
...: 'boosting_type': 'gbdt',
...: 'lambda_l1': trial.suggest_loguniform('lambda_l1', 1e-8, 10.0),
...: 'lambda_l2': trial.suggest_loguniform('lambda_l2', 1e-8, 10.0),
...: 'num_leaves': trial.suggest_int('num_leaves', 2, 256),
...: 'feature_fraction': trial.suggest_uniform('feature_fraction', 0.4, 1.0),
...: 'bagging_fraction': trial.suggest_uniform('bagging_fraction', 0.4, 1.0),
...: 'bagging_freq': trial.suggest_int('bagging_freq', 1, 7),
...: 'min_child_samples': trial.suggest_int('min_child_samples', 5, 100),
...: }
...:
...: gbm = lgb.train(param, dtrain)
...: preds = gbm.predict(test_x)
...: pred_labels = np.rint(preds)
...: accuracy = sklearn.metrics.accuracy_score(test_y, pred_labels)
...: return accuracy
...:
In [13]: study = optuna.create_study(direction='maximize')
...: study.optimize(objective, n_trials=100)
[I 2019-10-14 20:56:47,529] Finished trial#0 resulted in value: 0.951048951048951. Current best value is 0.951048951048951 with parameters: {'lambda_l1': 6.962619943277916e-08, 'lambda_l2': 2.4560252117247406e-05, 'num_leaves': 249, 'feature_fraction': 0.7647391158856618, 'bagging_fraction': 0.759593045720237, 'bagging_freq': 1, 'min_child_samples': 94}.
[I 2019-10-14 20:56:47,681] Finished trial#1 resulted in value: 0.9230769230769231. Current best value is 0.951048951048951 with parameters: {'lambda_l1': 6.962619943277916e-08, 'lambda_l2': 2.4560252117247406e-05, 'num_leaves': 249, 'feature_fraction': 0.7647391158856618, 'bagging_fraction': 0.759593045720237, 'bagging_freq': 1, 'min_child_samples': 94}.
[I 2019-10-14 20:56:47,815] Finished trial#2 resulted in value: 0.9790209790209791. Current best value is 0.9790209790209791 with parameters: {'lambda_l1': 0.009376054218409027, 'lambda_l2': 1.892546246456281, 'num_leaves': 208, 'feature_fraction': 0.40592732682242627, 'bagging_fraction': 0.9614938668620573, 'bagging_freq': 5, 'min_child_samples': 65}.
...
[I 2019-10-14 20:57:11,652] Finished trial#97 resulted in value: 0.965034965034965. Current best value is 1.0 with parameters: {'lambda_l1': 1.8269284172146948e-07, 'lambda_l2': 0.20632817551331567, 'num_leaves': 225, 'feature_fraction': 0.5131845782464036, 'bagging_fraction': 0.99436585113325, 'bagging_freq': 6, 'min_child_samples': 64}.
[I 2019-10-14 20:57:11,955] Finished trial#98 resulted in value: 0.986013986013986. Current best value is 1.0 with parameters: {'lambda_l1': 1.8269284172146948e-07, 'lambda_l2': 0.20632817551331567, 'num_leaves': 225, 'feature_fraction': 0.5131845782464036, 'bagging_fraction': 0.99436585113325, 'bagging_freq': 6, 'min_child_samples': 64}.
[I 2019-10-14 20:57:12,273] Finished trial#99 resulted in value: 0.972027972027972. Current best value is 1.0 with parameters: {'lambda_l1': 1.8269284172146948e-07, 'lambda_l2': 0.20632817551331567, 'num_leaves': 225, 'feature_fraction': 0.5131845782464036, 'bagging_fraction': 0.99436585113325, 'bagging_freq': 6, 'min_child_samples': 64}.
In [14]: print('Best trial:')
...: trial = study.best_trial
...:
...: print(' Value: {}'.format(trial.value))
...:
...: print(' Params: ')
...: for key, value in trial.params.items():
...: print(' {}: {}'.format(key, value))
...:
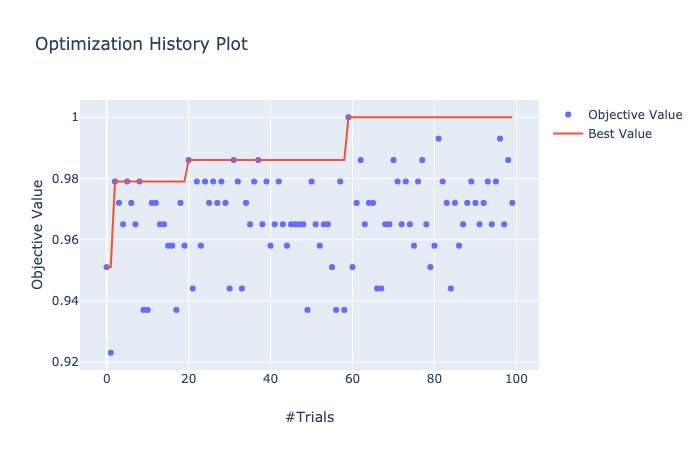
Best trial:
Value: 1.0
Params:
lambda_l1: 1.8269284172146948e-07
lambda_l2: 0.20632817551331567
num_leaves: 225
feature_fraction: 0.5131845782464036
bagging_fraction: 0.99436585113325
bagging_freq: 6
min_child_samples: 64
visualization.plot_optimization_history() で最適化の履歴を可視化できる。
In [15]: from optuna.visualization import plot_optimization_history
...: plot_optimization_history(study)
Pruning
Pruning (a.k.a automated early-stopping) により見込みの薄い trials を早期に終了することでパラメータ探索を効率化する。pfnet/optuna の examples/pruning/lightgbm_integration.py を実行する。
In [16]: def objective(trial):
...: data, target = sklearn.datasets.load_breast_cancer(return_X_y=True)
...: train_x, test_x, train_y, test_y = train_test_split(data, target, test_size=0.25)
...: dtrain = lgb.Dataset(train_x, label=train_y)
...: dtest = lgb.Dataset(test_x, label=test_y)
...:
...: param = {
...: 'objective': 'binary',
...: 'metric': 'auc',
...: 'verbosity': -1,
...: 'boosting_type': 'gbdt',
...: 'lambda_l1': trial.suggest_loguniform('lambda_l1', 1e-8, 10.0),
...: 'lambda_l2': trial.suggest_loguniform('lambda_l2', 1e-8, 10.0),
...: 'num_leaves': trial.suggest_int('num_leaves', 2, 256),
...: 'feature_fraction': trial.suggest_uniform('feature_fraction', 0.4, 1.0),
...: 'bagging_fraction': trial.suggest_uniform('bagging_fraction', 0.4, 1.0),
...: 'bagging_freq': trial.suggest_int('bagging_freq', 1, 7),
...: 'min_child_samples': trial.suggest_int('min_child_samples', 5, 100),
...: }
...:
...: # Add a callback for pruning.
...: pruning_callback = optuna.integration.LightGBMPruningCallback(trial, 'auc')
...: gbm = lgb.train(
...: param, dtrain, valid_sets=[dtest], verbose_eval=False, callbacks=[pruning_callback])
...:
...: preds = gbm.predict(test_x)
...: pred_labels = np.rint(preds)
...: accuracy = sklearn.metrics.accuracy_score(test_y, pred_labels)
...: return accuracy
In [17]: # optuna.logging.set_verbosity(optuna.logging.WARNING)
...: study = optuna.create_study(pruner=optuna.pruners.MedianPruner(n_warmup_steps=10),
...: direction='maximize')
In [18]: study.optimize(objective, n_trials=100)
...: trial = study.best_trial
[I 2019-10-22 11:55:29,817] Finished trial#0 resulted in value: 0.958041958041958. Current best value is 0.958041958041958 with parameters: {'lambda_l1': 1.9570352469571285e-07, 'lambda_l2': 1.1459013975087378, 'num_leaves': 34, 'feature_fraction': 0.8997956033930001, 'bagging_fraction': 0.7003740217574765, 'bagging_freq': 1, 'min_child_samples': 33}.
[I 2019-10-22 11:55:30,905] Finished trial#1 resulted in value: 0.972027972027972. Current best value is 0.972027972027972 with parameters: {'lambda_l1': 2.8042253461628537e-05, 'lambda_l2': 3.4975210119502136, 'num_leaves': 195, 'feature_fraction': 0.8203075154545054, 'bagging_fraction': 0.9364548026623444, 'bagging_freq': 2, 'min_child_samples': 7}.
[I 2019-10-22 11:55:31,443] Finished trial#2 resulted in value: 0.9370629370629371. Current best value is 0.972027972027972 with parameters: {'lambda_l1': 2.8042253461628537e-05, 'lambda_l2': 3.4975210119502136, 'num_leaves': 195, 'feature_fraction': 0.8203075154545054, 'bagging_fraction': 0.9364548026623444, 'bagging_freq': 2, 'min_child_samples': 7}.
...
[I 2019-10-22 11:55:51,815] Setting status of trial#22 as TrialState.PRUNED. Trial was pruned at iteration 11.
[I 2019-10-22 11:55:52,392] Setting status of trial#23 as TrialState.PRUNED. Trial was pruned at iteration 11.
[I 2019-10-22 11:55:52,952] Setting status of trial#24 as TrialState.PRUNED. Trial was pruned at iteration 11.
...
[I 2019-10-22 11:57:01,874] Finished trial#92 resulted in value: 0.965034965034965. Current best value is 1.0 with parameters: {'lambda_l1': 0.2876610835417842, 'lambda_l2': 0.000175639598247342, 'num_leaves': 56, 'feature_fraction': 0.9231284444098429, 'bagging_fraction': 0.718131583563206, 'bagging_freq': 4, 'min_child_samples': 84}.
...
[I 2019-10-22 11:57:05,871] Setting status of trial#97 as TrialState.PRUNED. Trial was pruned at iteration 11.
[I 2019-10-22 11:57:06,830] Setting status of trial#98 as TrialState.PRUNED. Trial was pruned at iteration 11.
[I 2019-10-22 11:57:07,610] Setting status of trial#99 as TrialState.PRUNED. Trial was pruned at iteration 11.
In [19]: print(' Value: {}'.format(trial.value))
...:
...: print(' Params: ')
...: for key, value in trial.params.items():
...: print(' {}: {}'.format(key, value))
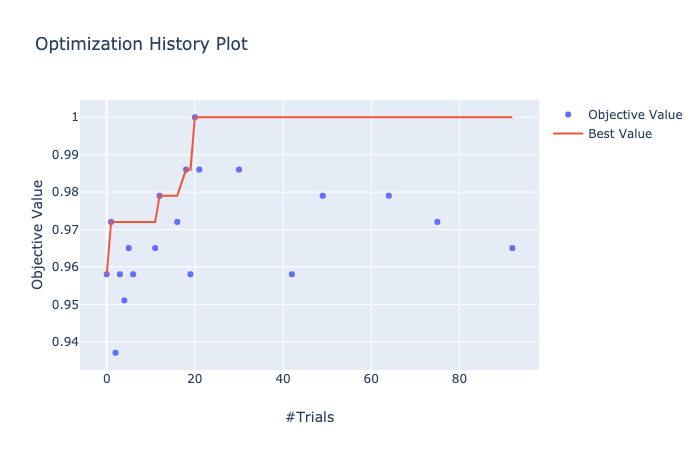
Value: 1.0
Params:
lambda_l1: 0.2876610835417842
lambda_l2: 0.000175639598247342
num_leaves: 56
feature_fraction: 0.9231284444098429
bagging_fraction: 0.718131583563206
bagging_freq: 4
min_child_samples: 84
先ほどと同様に visualization.plot_optimization_history() で最適化の履歴を可視化。
In [20]: from optuna.visualization import plot_optimization_history
...: plot_optimization_history(study)
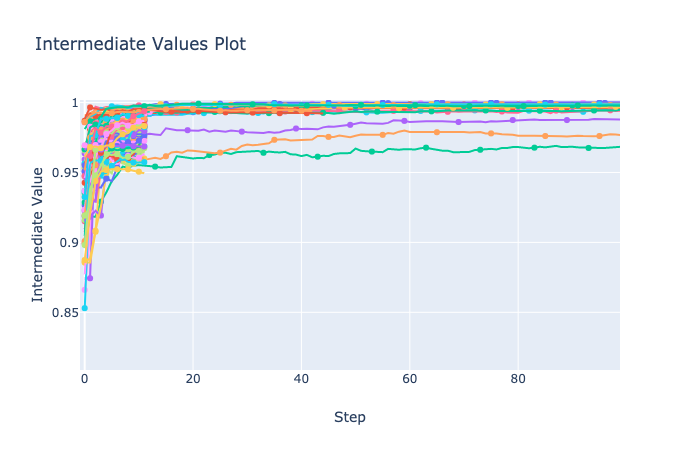
 visualization.plot_intermediate_values() で全ての trials の中間値を可視化できる。
visualization.plot_intermediate_values() で全ての trials の中間値を可視化できる。
In [21]: from optuna.visualization import plot_intermediate_values
...: plot_intermediate_values(study)n_warmup_steps で指定した 10 Steps で Pruning が行われていることが確認できる。
おわりに
今回は macOS に Optuna と LightGBM をインストールし pfnet/optuna に含まれる LightGBM の example を実行したところまでを備忘録として書きました。
[1] Optuna: A Next-generation Hyperparameter Optimization Framework
[2] 有名ライブラリと比較した LightGBM の現在
[3] 手を動かして GBDT を理解してみる
[4] 6.2.7. Breast cancer wisconsin (diagnostic) dataset
[5] 明治大学講演資料「機械学習と自動ハイパーパラメタ最適化」佐野正太郎
[6] Successive Halvingの性能解析